RNA Quality Control service | DNA Quality Control service
IntroductionThe facility offers a service to evaluate the quality and quantity of nucleic acids on a standalone basis or for use in a downstream service that we offer. This service will provide a concentration (ng/uL) and a size profile for either RNA or DNA. Nucleic Acid concentrations will be measured fluorometrically using a Qubit fluorometer. Samples submitted for standalone QC should be separate aliquots from those samples destined for use in a sequencing or genotyping service offered by the facility. Residual sample volume of samples for the QC service will be discarded following their use.
Sample Requirements
For either DNA QC or RNA QC a volume of 5 uL is required of the nucleic acid. The samples will be expanded with either nuclease-free water (RNA) or 10mM Tris (DNA) prior to being assayed. The concentration of the nucleic acid reported to the investigator will be calculated for the original sample concentration that was delivered to the facility. The required vial type for this service is 0.2 mL 8-strip tubes. All vials will need to be barcoded and delivered to the sample collection refrigerator or freezer located on the first floor of the R building.
Result Delivery
Both the individually measured concentrations and the size profile of each sample (including RQN score for RNA) will be delivered to the investigator electronically. Currently, the result delivery interface for this service is under construction, so results will be emailed directly by the laboratory technician to the submitter of the samples. The size profile for the samples will be in .pdf format.
Turnaround time
The targeted turnaround time for result delivery of this service is one week following sample barcoding and delivery.
RNA QC service
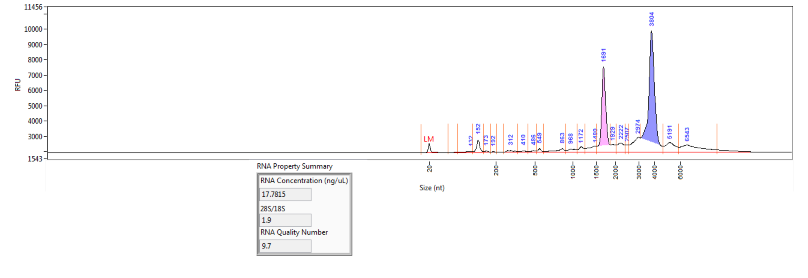
For the RNA QC service a RQN value is calculated for evaluation of RNA integrity. We utilize a Fragment Analyzer instrument from Advanced Analytical, which is similar to an Agilent BioAnalyzer in output. The RQN for RNA stands for RNA Quality Number, which is a similar metric to Agilent's RIN score. An additional metric, the DV200, can be evaluated specifically to determine the applicibility of degraded RNAs to NGS library preparation protocols.
DNA QC service
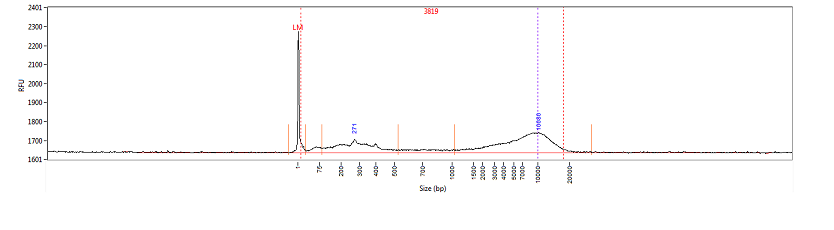
For the DNA QC service an average molecular weight of the DNA is returned. Both genomic DNA and NGS libraries can be evaluated. For genomic DNA a GQN metric can be evaluated to estimate quality. For NGS libraries, the percentage of 'clustergenic' library molecules can be estimated.
Pricing:
This service consists of two line items in CORES billing, A qubit concentration reading and an AATI size profile. Pricing is subject to change annually at the change of the fiscal year (July 1 to June 30).